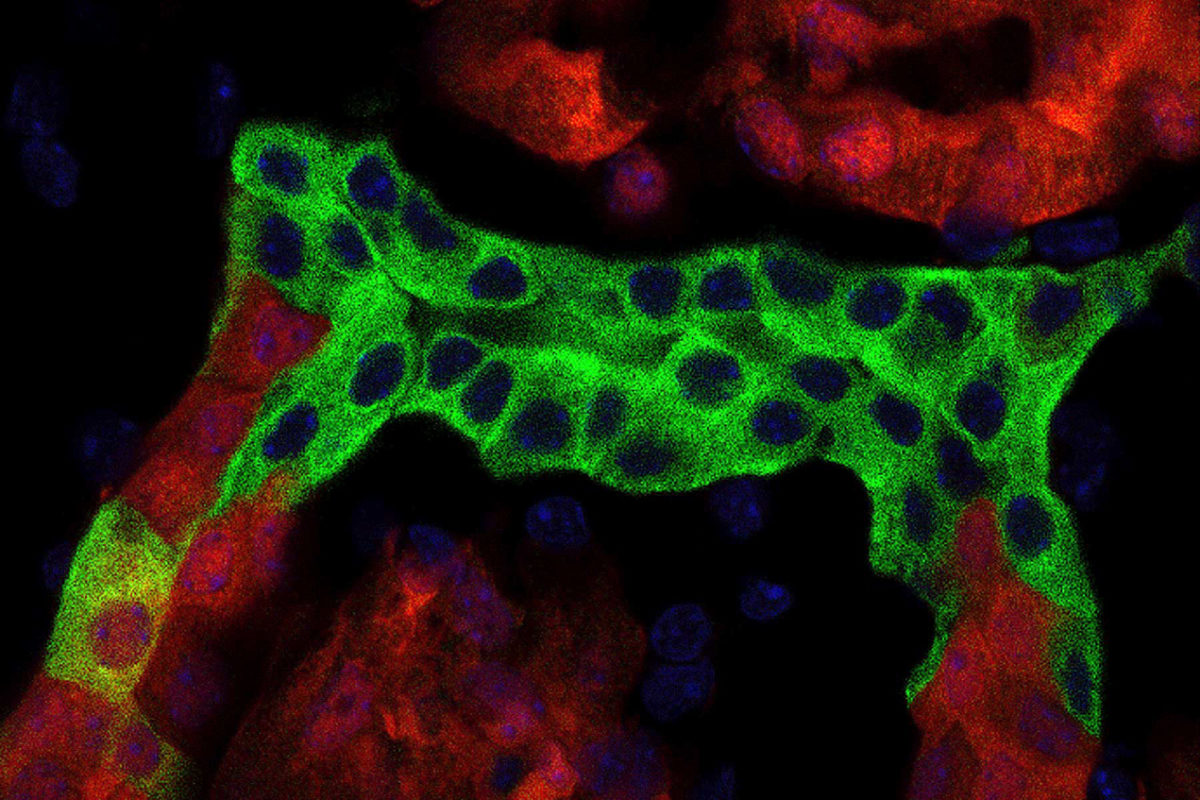
USC researchers have completed a detailed deconstruction of the kidney, revealing for the first time an intimate portrait of sexual differences and more in the organ.
The findings could benefit 37 million Americans suffering from kidney disease by helping to distinguish how it affects men and women differently. They can help doctors pinpoint genes linked to disease. And they can help scientists reprogram DNA to manipulate cells and to make new tissue lost to disease.
“What we’ve done is, for the first time, taken a complex organ to single-cell constituents, and then identified all the genetic activity occurring inside each cell, then reconstructed the kidney with a level of detail never seen before,” said Andrew McMahon, director of the Eli and Edythe Broad Center for Regenerative Medicine and Stem Cell Research at USC. ”Now, anyone can go to our online database and pinpoint the kidney cell types expressing any of the genes in the mammalian genome.”
The study was published today in Developmental Cell. McMahon is the lead author, joined by colleagues at the Keck School of Medicine of USC and at the University of Pennsylvania.
Comparing male and female mouse kidney cells
In the lab, the researchers focused on a mouse kidney, so certitude in translating findings to humans is unknown. Nonetheless, mouse organ-based research is proven to accelerate an understanding of human disease. And the structural similarities and sexual differences in key components of mammal kidneys — including mice, rats and humans — have been previously reported.
Kidney research is a priority for the USC Stem Cell initiative. Kidney disease affects 15% of U.S. adults, with notable differences in ethnic and sex-related susceptibility to kidney injury and disease. Every day, more than 240 people on dialysis die, according to the Centers for Disease Control and Prevention.
To deconstruct the kidney, McMahon and his team combined single-cell RNA sequencing of adult male and female mouse kidney cells and compared them to cell cultures in the lab. The result was a detailed, cell-by-cell look into the organ.
They found marked differences between sexes in the expression of genes associated with hormonal regulation, kidney disease and the kidney’s critical physiological activities. For example, they noticed differences between the sexes in the genes that code for enzymes that regulate blood pressure. The differences were especially evident in the proximal tubule region of the nephron, which is the workhorse tissue for reabsorption of essential factors such as glucose and metal ions, and the detoxification of drugs.
“These results highlight the need for a better understanding of sexual diversity within the human kidney,” McMahon said. “We know there are similarities between mice and humans in susceptibility to acute kidney injury — males are at a distinct disadvantage — and that sex differences can potentially impact drug studies and damage by kidney toxins.”
How sex can impact kidney disease and beyond
Indeed, the National Institutes of Health have emphasized that research needs to account for differences between sexes. Sex affects risk for disease, treatment and how people respond to medications. In the past, scientists studied male physiology and applied findings to women, so studies such as the new USC research underscore the importance of biological differences.
“Profound differences distinguish the male and female kidney,” McMahon said. “The kidney is the body’s regulator of fluid balance, and since women bear offspring, there are likely critical differences required in the mother for the benefit of both mother and offspring.”
The findings can benefit human health by improving an understanding of genetic programs that may influence drug trials, drug toxicity and cellular reprogramming, he said.
The searchable database housing this data, Kidney Cell Explorer, will enable researchers around the world to study gene expression in each cell in the correct anatomical framework for the kidney. McMahon said that clinicians who want to study a disease-associated gene can go to the database and see which cells express the gene, improving the linkage between genes, cells and disease.
The study authors include McMahon plus Andrew Ransick, Nils O. Lindström, Jing Liu, Jin-Jin Guo, Gregory F. Alvarado, Albert D. Kim and Hannah G. Black of USC; and Qin Zhu and Junhyong Kim of the University of Pennsylvania.
Work in McMahon’s lab was partially supported by National Institute of Health grants from the National Institute of Diabetes and Digestive and Kidney Diseases (NIDDK) (DK110792, DK054364, DK107350-01). Work by the Kim lab was supported in part by grants from the NIDDK (DK107350-02S1). Work by the team was also supported by a National Cancer Institute Cancer Center grant (P30CA014089). Both the McMahon and Kim labs are members of the NIDDK ReBuilding a Kidney Consortium. McMahon is also a member of the NIDDK Genitourinary Developmental Molecular Anatomy Project.
