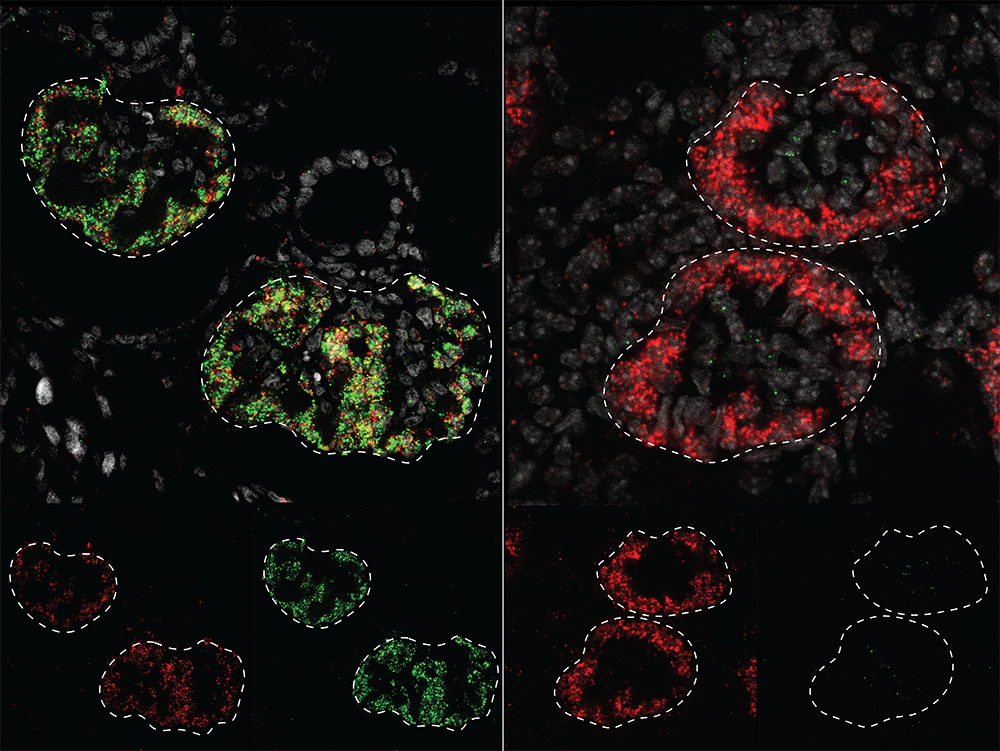
How similar is kidney development in humans and in the lab mice that form the foundation of basic medical research? In a new study published in Developmental Cell, USC Stem Cell scientists probe this question by comparing the activity and regulation of the genes that drive kidney development in lab mice and humans.
“While we do have a lot in common with lab mice, our evolutionary paths diverged around 80 million years ago,” said corresponding author Andy McMahon, director of the Eli and Edythe Broad Center for Regenerative Medicine and Stem Cell Research at USC, and W.M. Keck Provost and University Professor of Stem Cell Biology and Regenerative Medicine, and Biological Sciences. “On the anatomical level, there are obvious differences in mouse and human kidneys in terms of the overall organ size, number of filtering units, and patterning of the ducts and lobes. We wanted to deepen our understanding of these fundamental differences to the level of the underlying genes and gene regulators that orchestrate kidney development.”
To accomplish this, first author Sunghyun Kim in the McMahon lab worked with his colleagues to build atlases comparing gene activity and regulation in different cell types in developing mouse and human kidneys. The scientists could then pinpoint similarities and differences between the two species and identify cell type- and species-specific genetic and gene regulatory programs relevant to kidney development and disease.
Many genes, such as the one that encodes the molecule PCDH15, which helps cells adhere to each other, showed human-specific patterns of activity. These genes tended to be associated with cell interaction and migration, and might be necessary for building a complex, human-sized kidney during the relatively long period of embryonic development.
Other genes, including NTNG1, a gene normally associated with nerve cell development, may be used specifically in the human kidney to guide human developmental processes.
Many gene regulators were also human-specific. Some of these have been associated with chronic kidney disease or congenital anomalies of kidney and urinary tract.
“By identifying human-specific gene regulatory regions, we were able to link these to regions previously associated with kidney disease, showing a potential for our research to provide clinical insight,” said Kim, a recent PhD graduate from USC currently pursuing postdoctoral studies at the Massachusetts General Hospital in Boston.
Additional co-authors are Kari Koppitch, Riana K. Parvez, Jinjin Guo, MaryAnne Achieng, Jack Schnell, and Nils O. Lindström from USC.
The work was federally funded by the National Institute of Diabetes and Digestive and Kidney Diseases (grants R37DK054364 and UC2DK126024), and privately funded by the Chan Zuckerberg Initiative (grant WU-20-101) as part of the Seed Network of the Human Cell Atlas consortium.
Disclosure
McMahon is currently or has recently been a consultant or scientific advisor to Novartis, eGENESIS, Trestle Biotherapeutics, and IVIVA Medical. All authors declare that they have no conflicts of interest.
