The “secretome” refers to proteins that are secreted by a cell, a tissue or an organism. In a new study published in Open Biology, USC Stem Cell scientist Andy McMahon and his collaborators introduce an elegant new way to label and study the secretome in a living organism.
“The secretome orchestrates the subtle and complex processes of embryonic development, maintains the function of individual organs, and coordinates organ activity through inter-organ communication,” said McMahon, chair of the Department of Stem Cell Biology and Regenerative Medicine at USC. “However, it can be challenging to track which cells are secreting proteins, and which cells are being targeted.”
To address this challenge, co-first authors Rui Yang and Amanda S. Meyer at USC and Ilia A. Droujinine formerly at Harvard Medical School and now at Scripps Research generated mice with a built-in system for labeling and tracking the secretome.
“This built-in system is like a “passport system in the body, as we are identifying where proteins are coming from and where they’re going,” said Droujinine, who is a Scripps Research fellow and principal investigator in the Department of Molecular Medicine.
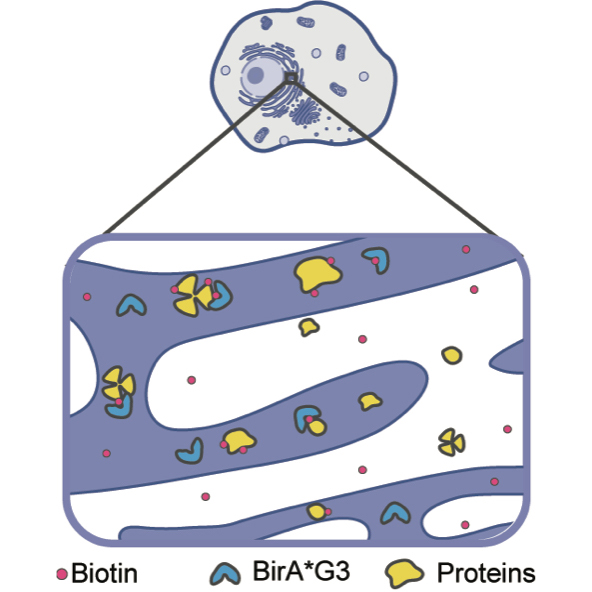
To establish this system, the research team started by genetically editing mouse embryonic stem cells to encode an enzyme engineered in Alice Ting’s laboratory at Stanford University. The enzyme, BirA*G3, indiscriminately tags nearby proteins with the vitamin B7 derivative, biotin. The reason that biotin is a useful tag is because it binds strongly the protein streptavidin, so that biotin-tagged proteins can be separated from other proteins in the cell and blood through a readily available technology called “streptavidin affinity purification.”
The next step was to ensure that BirA*G3 specifically tagged only the proteins of the secretome. These proteins originate in a structure within the cell known as the endoplasmic reticulum, or ER. Some of these proteins remain in the ER, while others are either secreted or incorporated into the cell’s surrounding membrane.
To specifically label only these proteins, BirA*G3 was itself tagged with four extra protein building blocks, or peptides. These four peptides, known as KDEL, served as a signal to the cell to retain BirA*G3 in the ER. So when biotin was added, BirA*G3 only added biotin labels to the proteins traveling through the ER.
Using these genetically edited stem cells, the team then produced transgenic mice. When these mice were given biotin, the scientists were able to demonstrate BirA*G3 labeling and identify the secretomes for a number of organs including the brain, liver and kidney. The scientists also identified proteins that were secreted anyway, even though they weren’t marked with the typical signal indicating the cell should secrete them.
These transgenic mice enable scientists to label and study specific cell types and organs, facilitating studies of inter-organ communication.
“Our new mouse model demonstrates enhanced labeling efficiency of the secreted and ER proteins in specific cell types, providing a valuable resource for mapping and profiling the secretome,” said Meyer, who is a PhD candidate in the McMahon Lab at the Eli and Edythe Broad Center for Regenerative Medicine and Stem Cell Research at USC.
Yang, a postdoctoral fellow in the McMahon Lab, added: “Our mouse model will facilitate a deeper understanding of the secretome’s critical role in development, as well as in healthy and diseased adult states.”
The mice are now in a public repository, so that other scientists can benefit from this new technology.
“Given the central role of key secreted proteins such as insulin, there is a great deal of interest in identifying novel secreted proteins,” said McMahon. “Genome studies are suggesting that many new proteins remain to be characterized. We’re looking forward to a deep dive into this area now that we have validated the technology.”
Additional co-authors include: Jinjin Guo and Jill A. McMahon from USC; Namrata D. Udeshi, Dominique K. Carey, Charles Xu, and Steven A. Carr from the Broad Institute of Harvard and MIT; Yanhui Hu and David Rocco from Harvard Medical School; Norbert Perrimon from Harvard Medical School and the Howard Hughes Medical Institute; Qiao Fang from the University of Toronto; Jihui Sha and James Wohlschlegel from UCLA; Shishang Qin from Peking University; and Alice Y. Ting from the Chan Zuckerberg Biohub and Stanford University.
The work was supported by federal funding from a National Institutes of Health Director’s Transformative Research Award (5R01DK121409).